2019-09-01 by Quick Biology Inc.
Brain organization at the regional, cellular level is the key to understand our cognitive abilities, what is disrupted in brain diseases. Single-cell RNA- profiling (scRNA-seq) can classify different cell types. Elucidating brain cell types is critical of how the neuron cell communicates with each other. However, dissociating living cells for scRNA-seq from human is difficult.
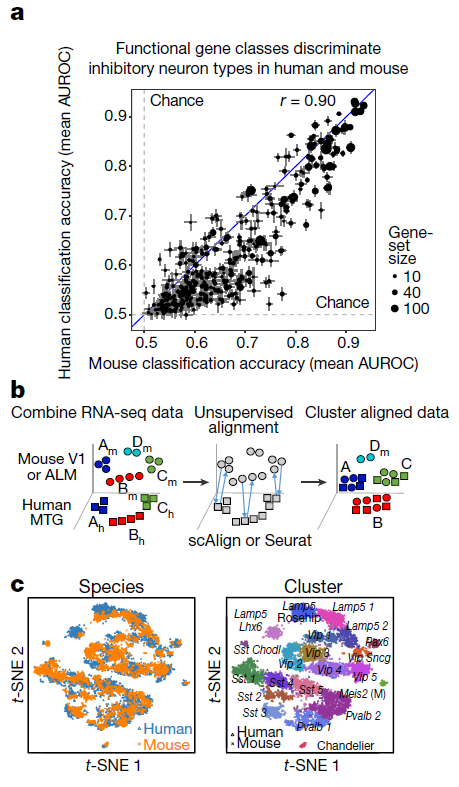
In recent publication Nature, Researchers Led by Dr. Ed Lein at Allen Institute for brain science (https://alleninstitute.org/what-we-do/brain-science/) used single-nucleus RNA-seq (snRNA-seq), making single cell transcriptome analysis from frozen brain tissue possible (Figure 1). By this method, they identified a highly diverse set of neuron types. More interestingly, by comparing to mouse brain transcriptome Atlas (single cell sequencing dataset), they revealed a surprisingly well-conversed cellular architecture (Figure 2). Their work provides us enhanced human brain cell resolution, accelerating our understanding of brain evolution.
For choosing whether single-nucleus or single-cell sequencing for your own research, please refer to Dr. Bosiljka Tasic and Dr. Ed S. Lein previous work (ref2).
Figure 1: Schematic of RNA-sequencing analysis of neuronal (NeuN+) nuclei isolated from human middle temporal gyrus (MTG). (ref1)
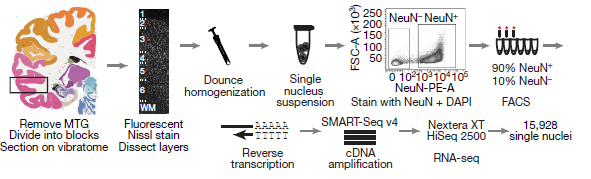
Figure 2: Evolutionary conservation of cell types between human and mouse. a, Similar functional gene families (n = 384 gene sets) discriminate inhibitory neuron types in human and mouse. Error bars correspond to the s.d. of mean MetaNeighbour AUROC scores across ten subsamples of cells. b, Schematic of unsupervised alignment and clustering of combined human (h) and mouse (m) cortical samples using scAlign or Seurat. c, t-SNE visualization of human (n = 3,594 nuclei) and mouse (n = 6,595 cells) inhibitory neuron clusters after alignment with scAlign. (ref1)
Quick Biology is an expert in NGS library preparation. Are you ready to sequence your samples of interest? Find More at Quick Biology.
Reference:
- 1. Hodge, R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019).
- 2. Bakken, T. E. et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One 13, 1–24 (2018).