2020-03-15 by Quick Biology Inc.
Transcriptome profiling using next generation sequencing has become routine in biological research. Typically, researchers do RNA extraction, poly(A) selection or ribosomal RNA depletion, first and second strand cDNA synthesis, sequencer adapter ligation, and final library PCR amplification (Fig. 1).
Bacterial transposase Tn5 is commonly used in double-stranded DNA tagmentation and ligating DNA ends to specific adaptors. Tn5 is the key reagent in conventional kits such as Illumina Nextera product line and Illumina mate-pair product line. In PNAS, researchers from China investigated that Tn5 also binds RNA/DNA heteroduplexes, can also add sequencing adapters onto RNA directly after reverse transcription (Fig. 2). Based on this newly discovered property of Tn5, they developed a new RNA-sequencing library preparation method, called SHERRY (Sequencing HEteRo RNA-DNA-hYbrid). As SHERRY gets rid of second stranded cDNA synthesis, links adapter sequencing by Tn5, not by A-tailing and T4 enzyme ligation, the entire library preparation time becomes shorter, initial RNA input requires less (Fig.3). SHERRY shows superior cross-sample robustness and comparable detectability for both bulk RNA and single cells compared with other commonly used methods (Fig.4).
Figure 1: Typical RNA-seq library preparation (PolyA selection Protocol) from Illumina.
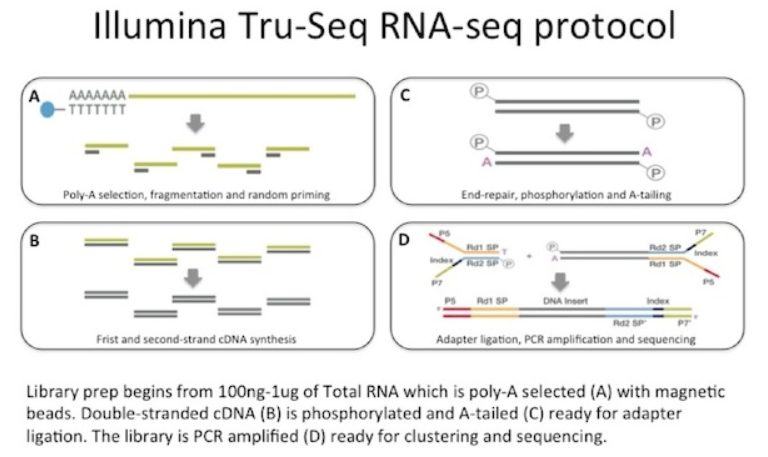
Figure 2: SHERRY Pilot Test. Verification of Tn5 tagmentation of RNA/DNA heteroduplexes. (A) Procedures of two ligation tests. Gray dotted box indicates negative results. (C) Strand test procedures (ref.1).
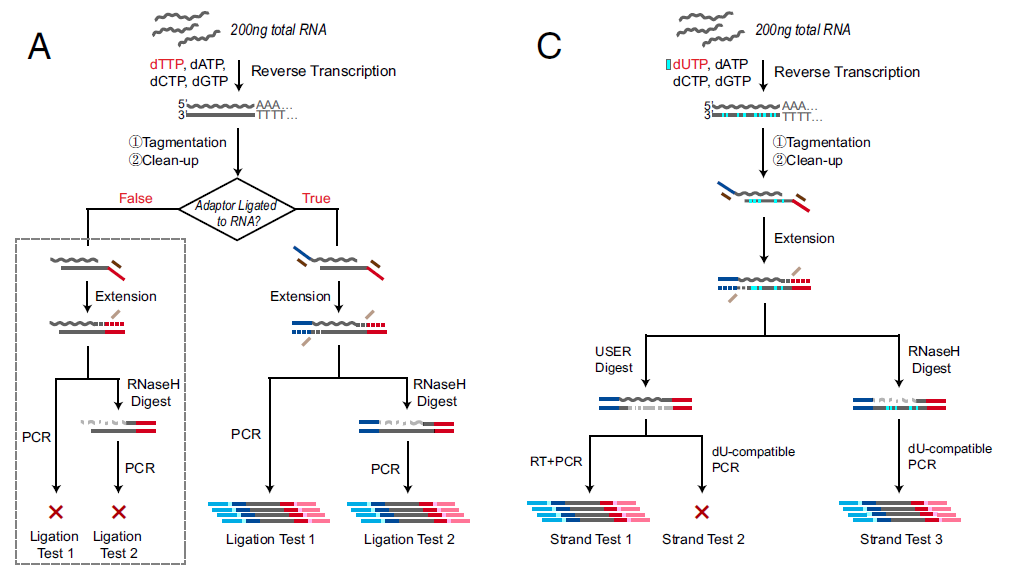
Figure 3: Workflow of SHERRY library preparation. Workflow of sequencing library preparation by SHERRY. The input can be a lysed single cell or extracted bulk RNA. After reverse transcription with oligo-dT primer, the hybrid is directly tagmented by Tn5, followed by gap-repair and enrichment PCR. Wavy and straight gray lines represent RNA and DNA, respectively. Dotted lines represent the track of the extension step.

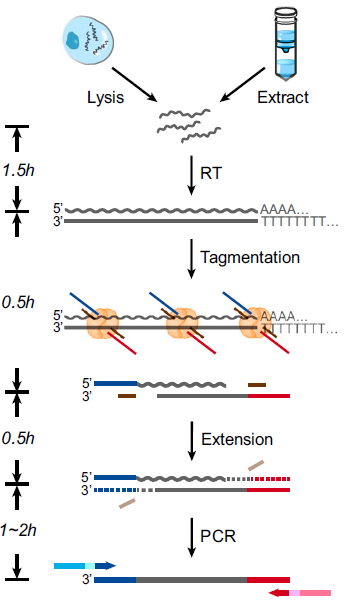
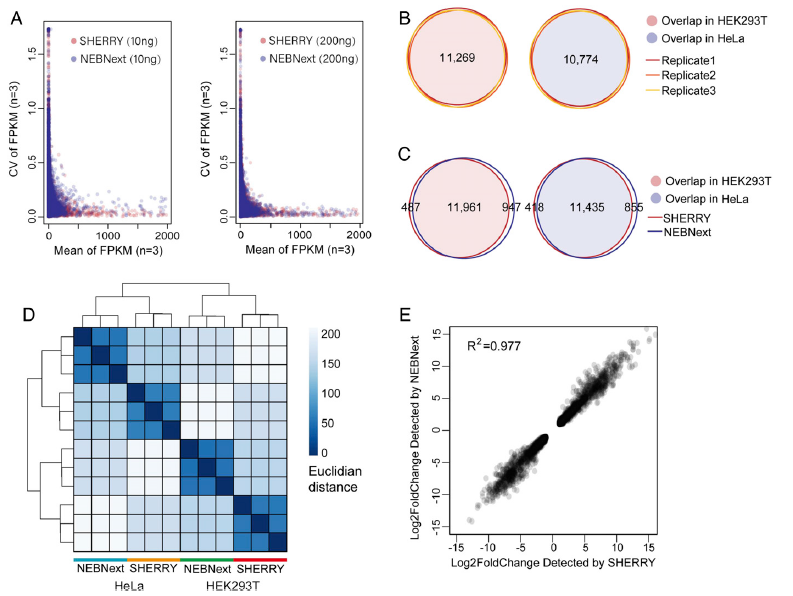
Figure 4: Comparison of SHERRY and currently on-the-market method (NEBNext). Performance of SHERRY with large RNA input. (A) Coefficient of variation (CV) across three replicates was plotted against the mean value of each gene’s FPKM (Fragments Per Kilobase of transcript per Million mapped reads). All experiments used HEK293T total RNA as input. (B) Genes detected by SHERRY in three replicates of 200 ng HEK293T or HeLa total RNA are plotted in Venn Diagrams. Numbers of common genes are indicated. (C) Common genes detected by SHERRY and NEBNext in the three replicates of 200 ng HEK293T or HeLa total RNA. (D) Distance heatmap of samples prepared by SHERRY or NEBNext for three replicates using 200 ng HEK293T or HeLa total RNA. The color bar indicates the Euclidian distance. (E) Correlation of gene expression fold-change identified by SHERRY and NEBNext. Involved genes are differentially expressed genes between HEK293T and HeLa detected by both methods (ref1).
Quick Biology is an expert in RNA-seq. Find More at Quick Biology.
Ref:
1. Di, L. et al. RNA sequencing by direct tagmentation of RNA/DNA hybrids. Proc. Natl. Acad. Sci. U. S. A. 117, 2886–2893 (2020).