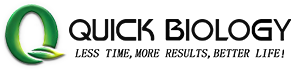2020-10-07 by Quick Biology
Since the first case of COVID-19 was reported in Dec 2019, a large amount of SARS-CoV-2 strains have been fully sequenced, the transcriptome is also being functionally annotated. Such an explosion of omics data of SARS-CoV2 has created a need for storing, analyzing, comparing these data. To well visualize, gain insights into SARS-CoV-2, Wang Lab at Washing University School of Medicine in St. Louis builds WashU Virus Genome Browser (https://virusgateway.wustl.edu/) (ref1, Figure 1); Haussler lab at UCSC Genomics Institute builds a similar interactive viewer for the SARS-CoV-2 into UCSC Genome browser (https://genome.ucsc.edu/covid19.html) (re2, Figure 2). The manuscripts describing how to use these web tools are published in Nature Genetics.
These comprehensive omics visualization resources offer but not limited to:
- 1. Genome organization and transcriptome features.
- 2. View sequence conservation with similar viruses and discover conserved immune-epitopes.
- 3. Guideline for PCR primer or CRISPR design for SARS-CoV-2 nucleic acid diagnostics.
- 4. Virus genome evolution, monitor, or track virus spreading.
Figure 1: Evaluating PCR primers for accumulating mutations with the WashU Virus Genome Browser. (ref1)
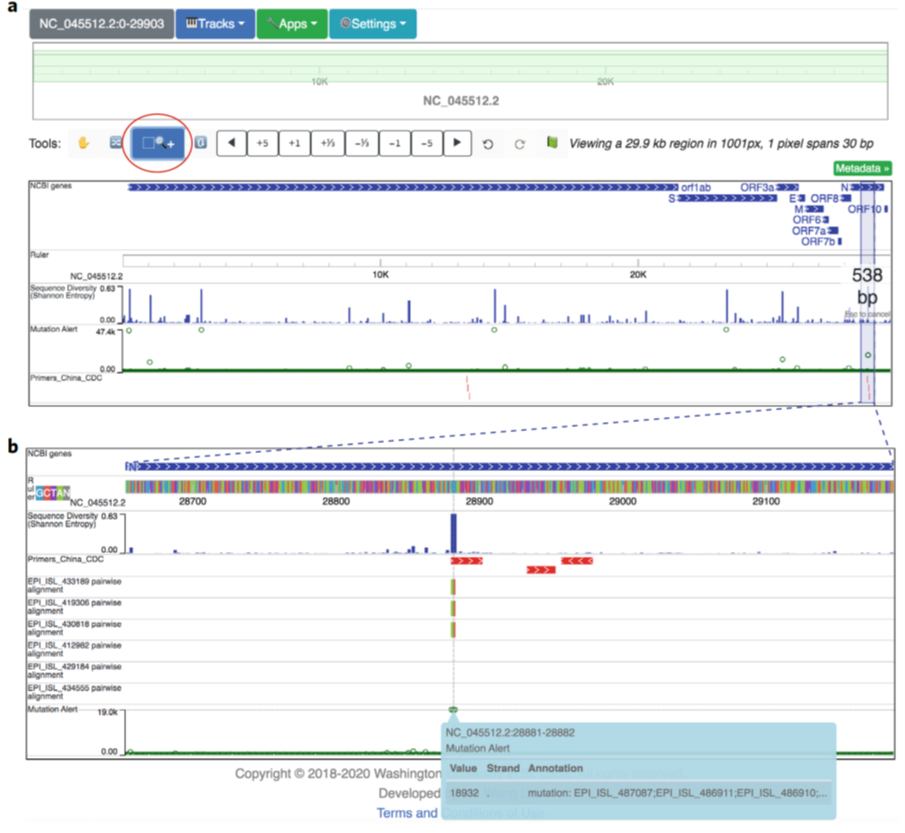
 Figure 2: UCSC Genome Browser user-interface structure. (ref2)
Figure 2: UCSC Genome Browser user-interface structure. (ref2)
Quick Biology has developed an extraction-free, laboratory-instrument-free for SARs-CoV2 test (Now in pre-EUA submission). Find More at Quick Biology.
Ref:
- 1. Flynn, J. A. et al. Exploring the coronavirus pandemic with the WashU Virus Genome Browser. Nat. Genet. 52, (2020).
- 2. Fernandes, J.D. et al. The UCSC SARS-CoV-2 Genome Browser. Nat. Genet. 52, (2020).