2020-07-20 by Quick Biology Inc.
Third-generation sequencing (also knowns as long-read sequencing) is a technology that reads the nucleotide sequences at single-molecule level. Comparing to existing methods (such as Illumina platform, BGI complete genomics platform, Thermofisher Ion Torrent platform, Qiagen GeneReader Platform) that infer nucleotide sequences by amplification and synthesis (SBS: sequencing by synthesis), the third-generation sequencing gives three major advantages:
- - Rapid sequencing pipeline. Sequencing does not base on synthesis, no clonal amplification step (which is for amplifying signal) in certain sequencers.
- - Improving de novo genome assembly, transcriptome gene isoform identification, fusion gene analysis, big-size genome structure variation analysis. All these benefits from the long reads.
- - Direct sequencing can identify base modifications in DNA and RNA.
Currently, there are two third-generation sequencing platforms, i.e. PacBio and Oxford Nanopore (see platform, data quality comparison in ref1, ref2). Here, we only introduce Oxford Nanopore. Nanopore platform works by monitoring changes to electrical current as nucleic acids are passed through a protein nanopore. By decoding electric changes, the researches can collect specific DNA or RNA sequences (Fig.1). Oxford Nanopore has more benefits in some applications comparing to PacBio.
- - Ultra-long to 2.3MB Vs. PacBio 10-15kb
- - Real-time and portable devices (Fig.2). The researcher can stop sequencing if they already get enough data; portable device make field deployable for nucleic acid detections, has been performed on board the ISS (International Space Station)
Figure 1: Illustration of the working principle of Nanopore sequencer (Adapted from ref 3). Inside the nanopore sequencer, an enzyme unwinds the DNA double helix and passes one of the strands through a nanopore – a microscopic channel in the center of a protein molecule sited in a membrane. A voltage is applied across the nanopore, causing ions in the solution to move through the pore and generate an ionic current. The channel is just the right size for the DNA strand to pass through. Since each of the four DNA bases is different in size, it blocks a different proportion of the nanopore as it passes through. This means there is more or less space for ions to pass through the nanopore, and so more or less current can flow across the gap. The ionic current is measured and the data is transferred to the computer and analyzed by specialist software that converts the measurements in real-time into the DNA sequence – a text file with the letters A, T, C, and G.
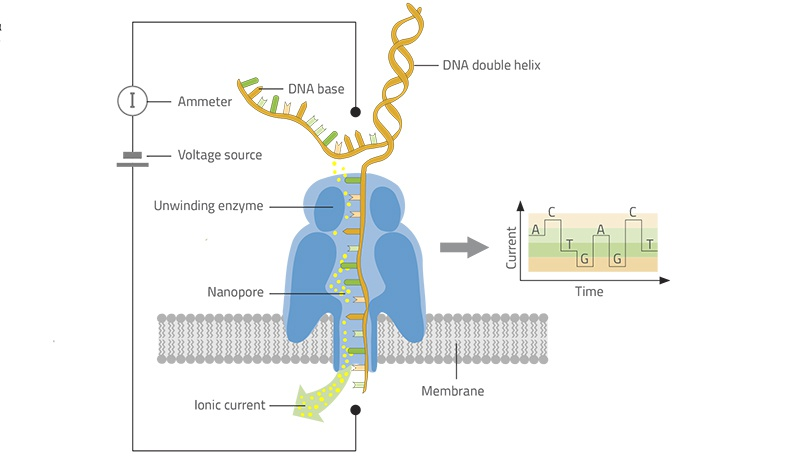
Figure 2: Portable Devices from Oxford Nanopore. (image from ref 4)

Nanopore sequencing offers advantages in all areas of research. Our offering includes DNA sequencing, as well as RNA and gene expression analysis and future technology for analyzing proteins.
Given the more requests and demonstrated utility of long reads sequencing, Quick Biology Inc. will start to provide Nanopore sequencing this Fall 2020. Find More at Quick Biology.
Ref:
- 1. Genehub Blog by Bhagyashree Birla 2017: https://blog.genohub.com/2017/06/16/pacbio-vs-oxford-nanopore-sequencing/
- 2. F1000Research Article from Dr. Au Lab: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5553090/pdf/f1000research-6-12836.pdf
- 3. Decoding DNA with a pocket-sized sequencer: https://www.scienceinschool.org/content/decoding-dna-pocket-sized-sequencer
- 4. Oxford Nanopore product: https://nanoporetech.com/applications