2020-08-03 by Quick Biology Inc.
The nucleus of the human cell harbors very densely packed chromosomes (Fig.1). Determining how these chromosomes are folded and interacted is critical in understanding gene regulation. Besides imaging approaches like FISH (fluorescence in situ hybridization), approaches based on chromosome conformation capture (3C), especially Hi-C (high-throughput chromosome conformation capture) is becoming more popular in current years (Fig. 2). In the HiC protocol, the cell firstly will be crosslinked in interacting loci; After digestion with restriction enzymes, the free end DNA molecules will be labeled by Biotin; Followed by ligation, purification, shearing and streptavidin beads pull down (Fig 3). Combining with next generation high-throughput sequencing, the HiC method is the first unbiased and genome-wide adaptation of 3C, providing a true all-by-all genome-wide interaction map (see comparisons of 3C based methods for detecting chromatin contacts in Table 1). Quick Biology is an expert in NGS assay development, our scientists have finished the internal test, compared multiple current HiC commercial kits, and optimized the HiC protocol. We are open, and now ready to help you with your HiC projects. Find out more at Quick Biology.
Figure 1: Chromatin organization across genomic scales. (source from ref 1). The chromatin fiber from one chromosome is unraveled to illustrate four different organization levels. Chromatin conformations are presented from low (top) to high (bottom) resolutions. The chromatin fiber and corresponding chromosome territory are shown in pink. A and B compartments (multi-megabase scale) are shown separately to highlight their inherently distinct nature, although there is no evidence that their conformations differ at the level of TADs (megabase scale). Three examples of chromatin looping (sub-megabase scale) are shown: (i) enhancer-promoter, (ii) enhancer-silencer, and (iii) insulator-insulator. E, enhancer; P, promoter; S, silencer; I, insulator.
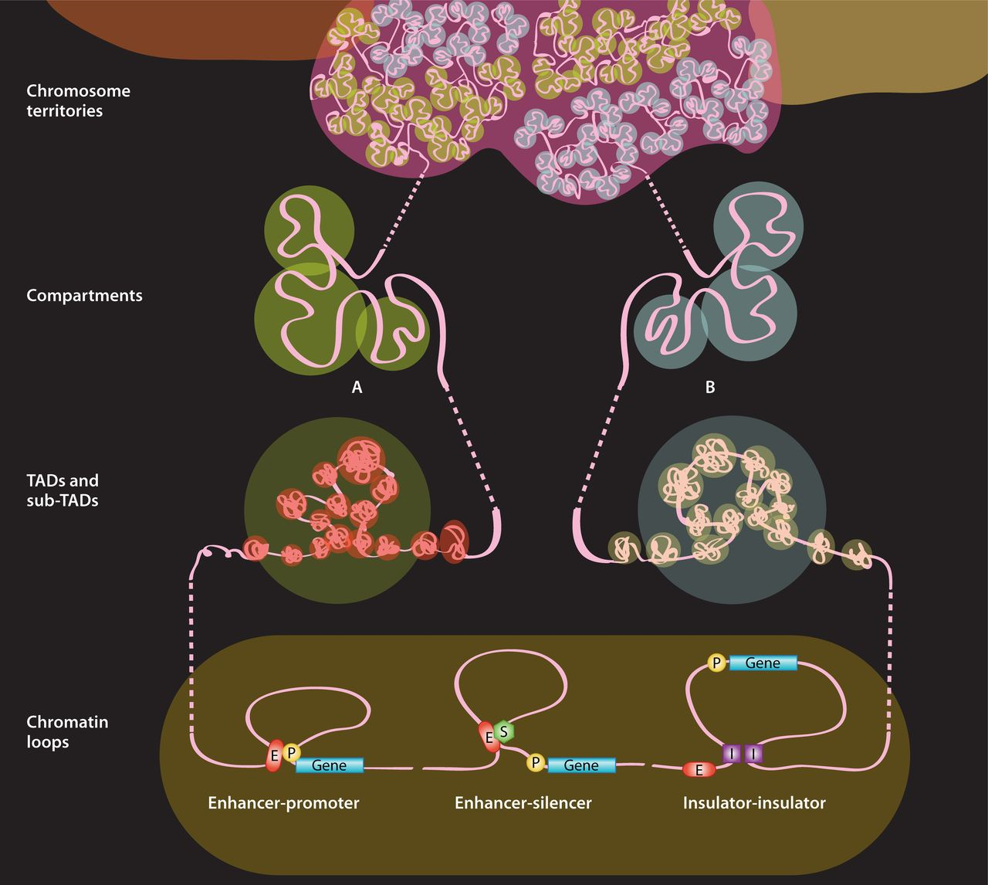
Figure 2: Demonstration of chromosome conformation capture-based methods (from ref 2). In chromosome conformation capture (3C)‑based methods (see panel a of the figure), cells are crosslinked with formaldehyde to link chromatin segments covalently that are in close spatial proximity. Next, chromatin is fragmented by restriction digestion or sonication. Crosslinked fragments are then ligated to form unique hybrid DNA molecules. Finally, the DNA is purified and analysed. The different 3C‑based methods differ only in the way that hybrid DNA molecules, each corresponding to an interaction event of a pair of loci, are detected and quantified (see panel b of the figure). In classical 3C experiments, single ligation products are detected by PCR one at the time using locus-specific primers. 4C (also known as ‘circular 3C’ or ‘3C‑on‑chip’) uses inverse PCR to generate genome-wide interaction profiles for single loci. 5C combines 3C with hybrid capture approaches to identify up to millions of interactions in parallel between two large sets of loci. chromatin interaction analysis by paired-end tag sequencing (ChIA–PET) method allows for genome-wide analysis of long-range interactions between sites bound by a protein of interest. Hi‑C provides a true all‑by‑all genome-wide interaction map, but the resolution of this map depends on the depth of sequencing. When several hundred million read pairs are obtained, as is currently routine, chromatin interactions in the mouse or human genome can be detected at 100 kb resolution.
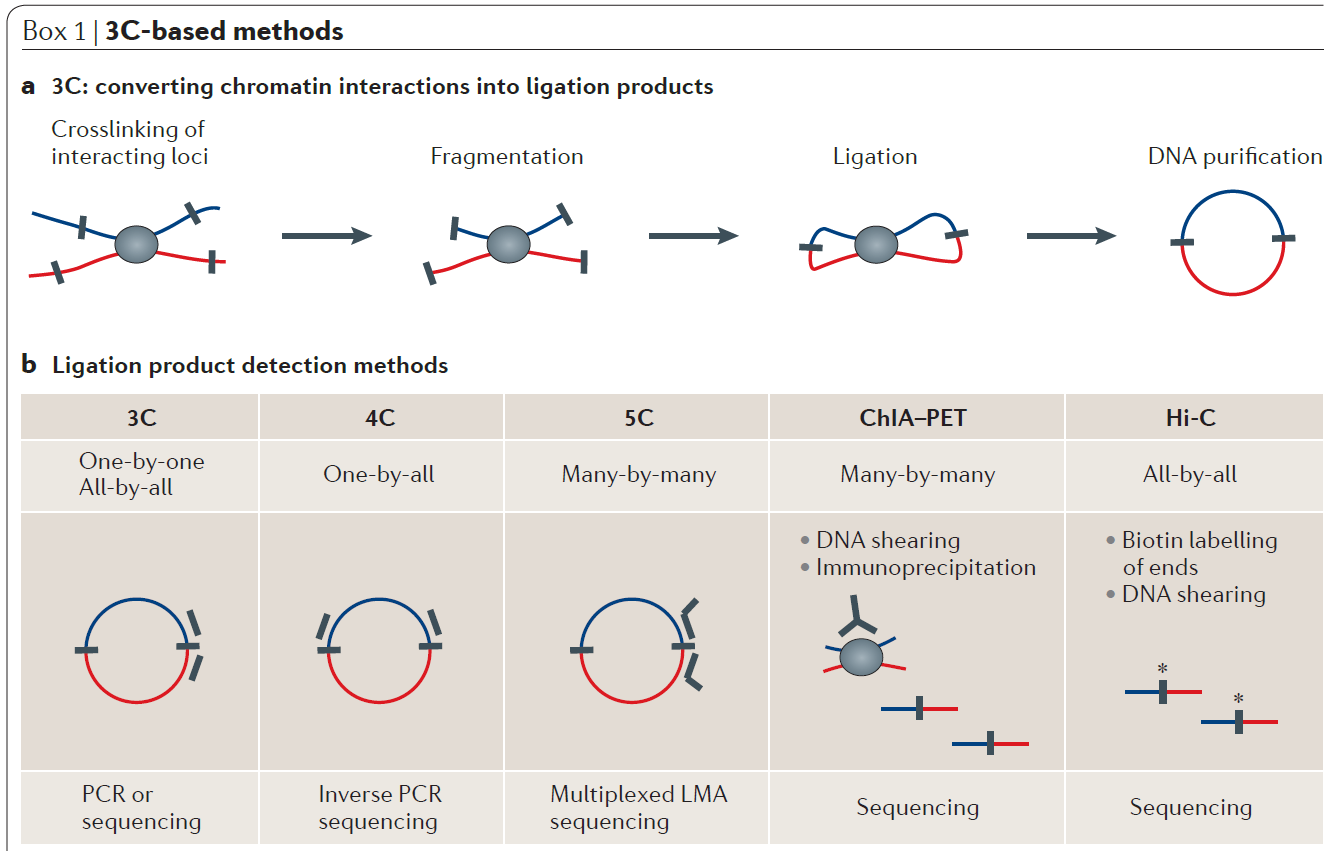
Figure 3: Quick Biology Hi-C workflow (from Arima-HiC user guide, ref 3)

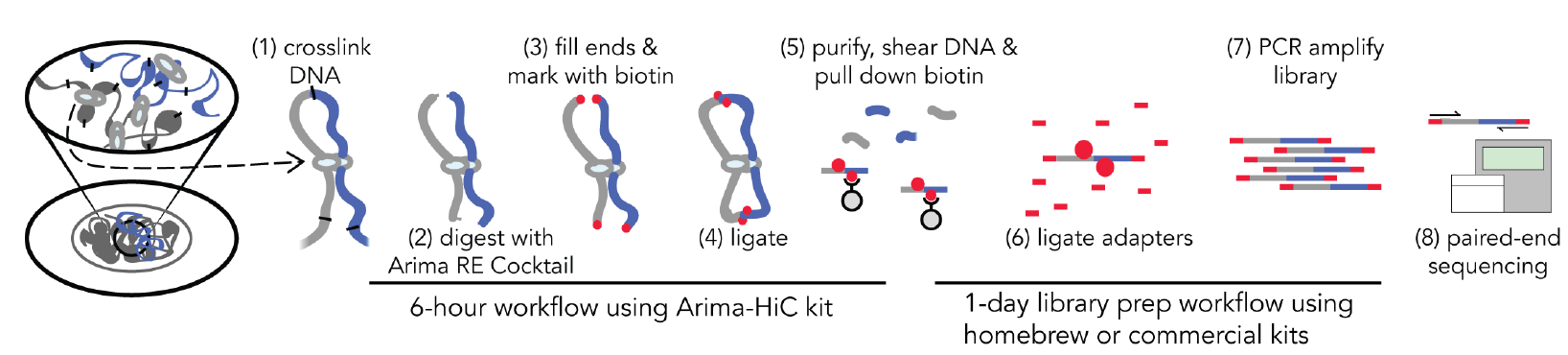
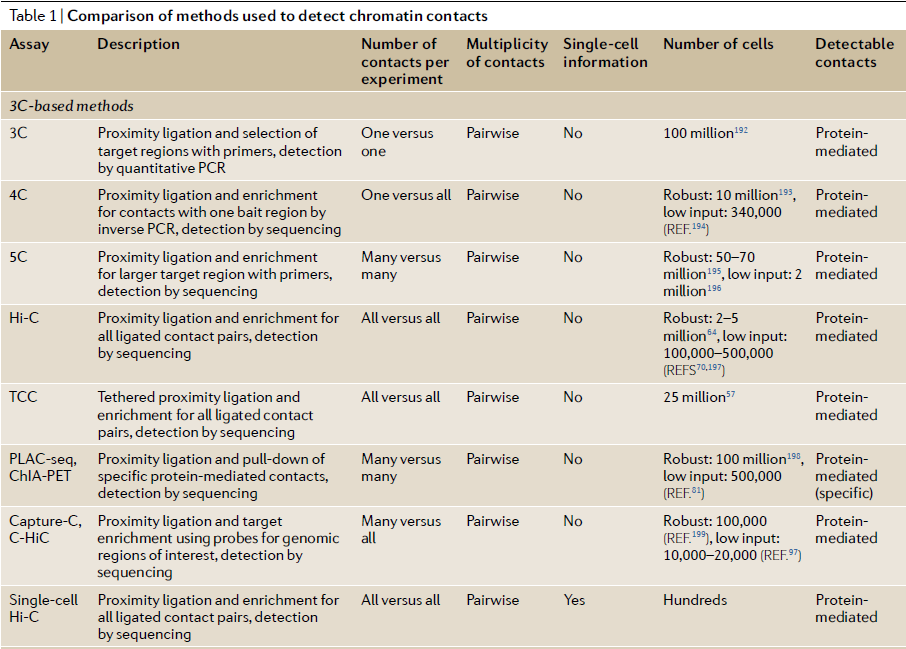
Table 1: Comparison of 3C methods to detect chromatin contacts (ref 4). Quick Biology is now offering both HiC and Capture-HiC services.
Ref:
- 1. James Fraser et al. An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol. Mol. Biol. Rev. 2015; doi:10.1128/MMBR.00006-15
- 2. Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
- 3. Arima Genomics: https://arimagenomics.com/kit
- 4. Kempfer, R. & Pombo, A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 21, 207–226 (2020).