2020-11-17 By Quick Biology
In recent years, single-cell transcriptome sequencing technology has provided powerful tools for solving key biomedical problems. Although the single-cell transcriptome solves the problem of cell heterogeneity, it cannot restore its specific spatial location, and it is impossible to accurately explore cell functions. Spatial transcriptomics can fill this gap. Using spatial transcriptome technology, researchers can locate the gene expression information of cells in the tissue to the original spatial position of the tissue, thereby analyzing the differences in the function and gene expression of different parts of the tissue.
But the technology of spatial transcriptome is not yet mature. The existing detection methods have high technical requirements, including single-molecule fluorescence imaging systems that require high sensitivity, complex image analysis processes, and lengthy repetitive imaging workflows, which make it difficult to achieve large-scale research applications. In addition, most methods are based on limited probe hybridization techniques to detect known gene sequences, which limits the possibility of discovering new and mutated genes.
To overcome the above bottlenecks, the team of Professor Rong Fan from Yale University developed a spatial omics sequencing technology called DBiT-seq (Deterministic Barcoding in Tissue for spatial omics sequencing). This technology does not require complex imaging technology but uses high-throughput sequencing NGS and DNA barcode technology to obtain transcriptome and proteomics information while obtaining spatial information, which can achieve higher sample throughput and cost-effectiveness. By applying this technology to the analysis of mouse embryos, the results show that DBiT-seq can successfully capture the main tissue types in early organ formation in mice, as well as the microvascular endothelial cells and retinal pigment epithelial cells in the brain. Recently, the results of the study were published on Cell with the article titled "High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue" (Ref 1).
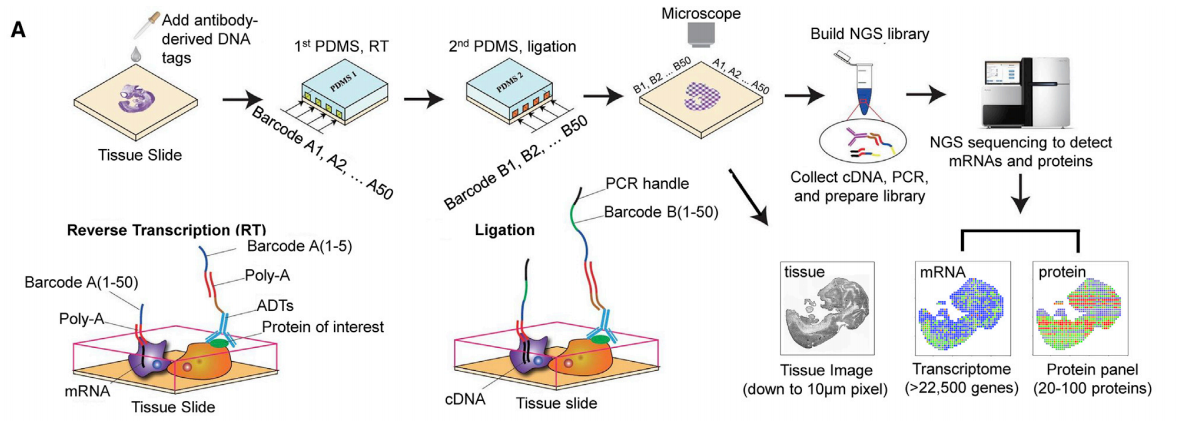
According to the article, DBiT-seq does not require tissue lysis to release mRNA and is compatible with existing formaldehyde-fixed tissue slides. This technology can simultaneously obtain mRNA and protein information in tissue slides through NGS sequencing. DBiT-seq can directly use a microfluidic chip containing 50 parallel microchannels to introduce DNA barcodes A and B into tissue section samples. This technology first synthesizes the first-strand cDNA in situ by reverse transcription, then the cDNA strand is incorporated into barcode A, and then DNA barcode B can be introduced.
After the above two steps, the tissue slices are connected in situ at the intersection through two sets of barcode systems to form an integrated point of tissue pixels. Each pixel point contains a different combination of barcodes A and B to form a two-dimensional image. In addition, to detect the proteome and transcriptome at the same time, tissue sections were also stained with 22 antibody-derived DNA tags (ADTs). Finally, after forming a spatial barcode combination, cDNAs are collected for PCR amplification, a sequencing library is constructed, and paired-end sequencing is performed using NGS (Figure 1).
Figure 1: DBiT-seq workflow (from Ref 1)
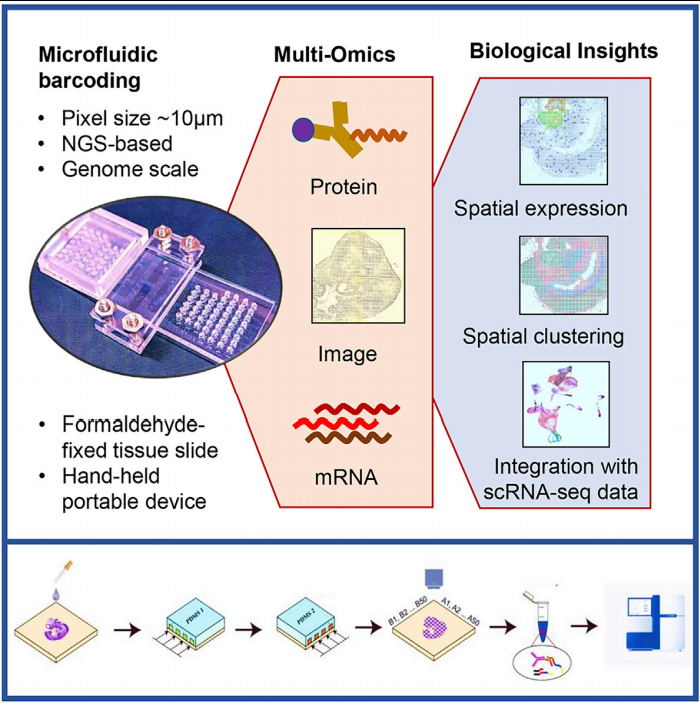
In summary, DBiT-seq technology can obtain transcriptome and proteomics information of tissue sections with high spatial resolution. This NGS-based detection method is unbiased and can simultaneously detect the entire transcriptome level, so it can be used to map genes in the tissue environment. In addition, DBiT-seq is fundamentally different from other spatial transcriptome-based methods and only requires a set of reagents and a simple device to perform experiments. Therefore, it has a strong application potential and can be applied in a wide range of biology and biomedical research, including developmental biology. Science, neuroscience, cancer, immunobiology and clinical pathology (Figure 2).
Figure 2: DBiT-seq potential applications (from Ref 1)
Quick Biology Inc. is very experienced in applying single-cell technology using NGS. Please find more information here.
Reference:
1. Liu et al., High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue, Cell (2020). Published online Nov 13, 2020.